How to train a decision tree in R?
Let's say I have the following datasets.
set.seed(123)
n <- nrow(iris)
indices <- sample(n, n * 0.8)
treino <- iris[indices, ]
teste <- iris[-indices, ]
How could I use R to train a decision tree capable of predicting the plant species based on its measurements?
It would be good if the answer included a brief explanation of what a decision tree is and the problems where this solution is often used.
1 answers
Without going too deep into the theoretical part, a classification tree is a mathematical model that uses the decision tree structure to classify data. Better than explaining this in words is to see the algorithm in action:
library(rpart)
library(rpart.plot)
modelo <- rpart(Species ~ ., method="class", data=iris)
prp(modelo, extra=1)
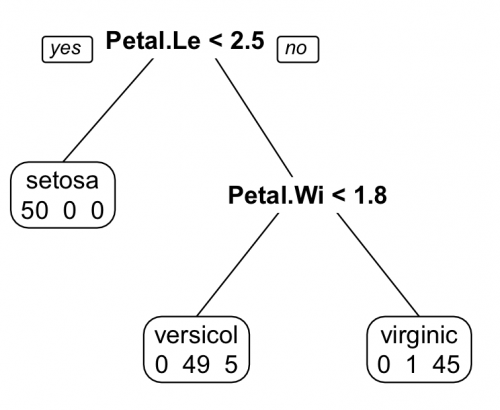
Note that a question is asked at each node in the tree. The answer to the question determines whether another question will be asked or whether the tree has come to an end and the sorting has been finished. In addition, the error of classification is determined at the end of the tree.
In this example, the questions the algorithm asked start with: Petal.Length < 2.5? If true, then the species is classified as setosa. Otherwise, another question is asked: Petal.Width < 1.8? If true, the species is classified as versicolor. Otherwise, it is classified as virginica.
Also notice that at the bottom of each end node of the tree are numbers that indicate the result of the sorting. At the first node, all 50 setosa were classified correctly. In the other two nodes, there were classification errors: of the 50 versicolor, 49 were classified correctly, but 5 were considered virginica. Of the 50 virginica, 45 were classified correctly and 1 was considered versicolor.
Note that in the original question, the author put the following code snippet:
set.seed(123)
n <- nrow(iris)
indices <- sample(n, n * 0.8)
treino <- iris[indices, ]
teste <- iris[-indices, ]
He did this because a problem that arises in fitting classification models is the over-fitting. The over-adjustment it occurs when the model fits the data very well, making it ineffective to predict new observations.
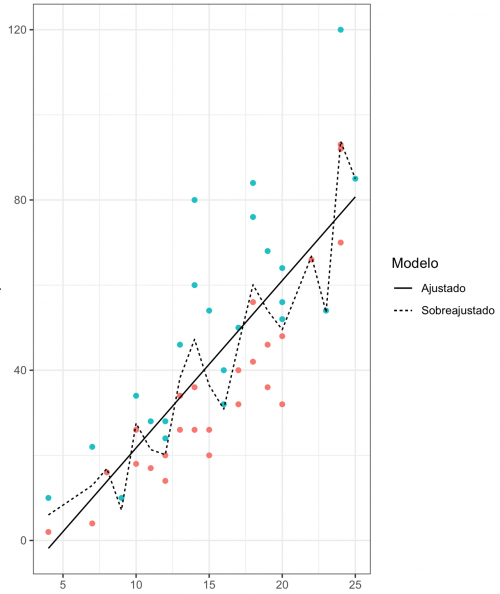
The graph below illustrates this concept in the context of a classification model.
There are two lines that separate the green dots from the red ones. Although the dashed line faithfully hits all the classifications it makes, it ends up not serving to generalize the result. If new observations enter the system, the model generated by dashed line will not have good predictive power and will not serve to classify new data. That is, it is very good, but only for a specific set of data. Therefore, the model defined by the continuous line, despite making more mistakes, ends up becoming more useful.
One way to avoid this problem is by randomly dividing the original dataset into two mutually exclusive parts. One of these parts is called a training set, and the other-a test set. The idea by behind this is to adjust the model to the training data and simulate the input of new observations through the test set. Thus, it is possible to check how well or how poorly the adjusted model is behaving when predicting observations that were not used in its adjustment.
This division is possible to be done the way it was done in the original post. In particular, I prefer to use a function called createDataPartition, present in the package 'caret'
library(caret)
# definir 75% dos dados para treino, 25% para teste
set.seed(1234)
trainIndex <- createDataPartition(iris$Species, p=0.75, list=FALSE)
iris_treino <- iris[ trainIndex, ]
iris_teste <- iris[-trainIndex, ]
When doing iris_treino <- iris[ trainIndex, ] I I'm saying that the dataset iris_treino will have the rows of iris with the numbers present in trainIndex. Similarly, iris_teste <- iris[-trainIndex, ] says that the dataset iris_teste not will have the lines of iris with the numbers present in trainIndex
I now have two new data frames on my desktop. 75% of observations are in the training set and 25% in the test set. This division is arbitrary. Normally, it is recommended that the training set have 70% to 80% of the remarks. The rest of the observations will be part of the test set.
It happens that this division of data into two groups has a disadvantage. This ends up causing us to have less data to adjust the model. And with less data to fit the model, less information we have. With less information, the worse our model will look. One way to reduce this effect is through cross-validation.
A cross-validation is another method used to avoid over-fit on Model. The idea is to fit the same model several times into partitions (mutually exclusive sets) of the original training set. In this example I will use a method called cross-validation fol k $ - fold.
This technique consists of five steps:
-
Separate the training set into k folds (or partitions)
Set the model to k-1 folds
-
Test the model in the remaining fold
-
Repeat Steps 2 and 3 until all folds have been used for testing
-
Calculate model accuracy
However, we need to define the number of folds to be used in cross-validation. In general, the literature suggests that from 5 to 10 folds be used. The performance of algorithms does not it improves considerably if we greatly increase the number of folds.
Depending on the size of the dataset, it is possible that many folds end up leaving us with no observations for the test sets within the cross-validation. For this reason, it is always good to control this parameter according to the dataset we are studying.
With these defined techniques, we can finally move on to model tuning.
O caret uses two functions to fit models to data, called train and trainControl. Basically, the function trainControl sets the parameters used in the model fit. Below I am exemplifying how to define that we want to do cross-validation with 5 folds.
fitControl <- trainControl(method = "cv",
number = 5)
With the parameters of the cross-validation defined, we can leave for the adjustment itself.
ajuste_iris <- train(Species ~ .,
data = iris_treino,
method = "rpart",
trControl = fitControl)
The result of the adjustment is as follows:
CART
114 samples
4 predictor
3 classes: 'setosa', 'versicolor', 'virginica'
No pre-processing
Resampling: Cross-Validated (5 fold)
Summary of sample sizes: 92, 91, 90, 91, 92
Resampling results across tuning parameters:
cp Accuracy Kappa
0.0000000 0.9731225 0.9598083
0.4736842 0.7032938 0.5617284
0.5000000 0.4429513 0.1866667
Accuracy was used to select the optimal model using the largest value.
The final value used for the model was cp = 0.
I will not enter into the merit of what means the hyperparameter cp. What matters in this is result are Accuracy (accuracy) and Kappa (type the accuracy, except that this measure is normalized by the random chance of classification). I will leave a more detailed explanation of accuracy and Kappa open, but just know that both accuracy and Kappa vary between 0 and 1. However, the higher these values, the better the model fit.
Therefore, the adjusted model achieved an overall accuracy of 95% in the training data. But that's only half the work. We need to see how the model behaves in the test data. For this we will use the following commands:
predicao <- predict(ajuste_iris, iris_teste)
confusionMatrix(predicao, iris_teste$Species)
Confusion Matrix and Statistics
Reference
Prediction setosa versicolor virginica
setosa 12 0 0
versicolor 0 10 2
virginica 0 2 10
Overall Statistics
Accuracy : 0.8889
95% CI : (0.7394, 0.9689)
No Information Rate : 0.3333
P-Value [Acc > NIR] : 6.677e-12
Kappa : 0.8333
Mcnemar's Test P-Value : NA
Statistics by Class:
Class: setosa Class: versicolor Class: virginica
Sensitivity 1.0000 0.8333 0.8333
Specificity 1.0000 0.9167 0.9167
Pos Pred Value 1.0000 0.8333 0.8333
Neg Pred Value 1.0000 0.9167 0.9167
Prevalence 0.3333 0.3333 0.3333
Detection Rate 0.3333 0.2778 0.2778
Detection Prevalence 0.3333 0.3333 0.3333
Balanced Accuracy 1.0000 0.8750 0.8750
We have an accuracy of 88.89% in the test data. Not bad, considering we work with a small sample. In addition, the confusionMatrix command gives us other measures of fit quality, such as sensitivity (false negative rate) and specificity (false positive rate).